Doing so introduces Gibb’s phenomenon, as explained in: Projection and interpolation — FEniCS Tutorial @ Sorbonne
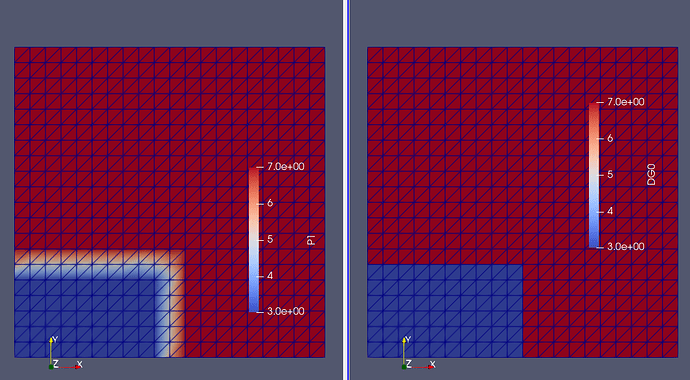
What you could do is to take the average from all cells associated with a given vertex, as it would not introduce oscillations as Gibb’s.
To do so I would loop through each vertex and find the connecting cells for each vertex and extract the values, i.e.
from mpi4py import MPI
import numpy as np
import dolfinx
mesh = dolfinx.mesh.create_unit_square(MPI.COMM_WORLD, 20, 20)
V = dolfinx.fem.functionspace(mesh, ("Lagrange", 1))
Q = dolfinx.fem.functionspace(mesh, ("DG", 0))
q = dolfinx.fem.Function(Q)
q.name = "DG0"
# Mark some of the cells with 3, the others with 7
left_cells = dolfinx.mesh.locate_entities(mesh, mesh.topology.dim, lambda x: (x[0] <= 0.5+1e-14) & (x[1] <= 0.3+1e-14))
q.x.array[:] = 7
q.interpolate(lambda x: np.full(x.shape[1], 3.0), cells=left_cells)
q.x.scatter_forward()
num_vertices = mesh.topology.index_map(
0).size_local + mesh.topology.index_map(0).num_ghosts
mesh.topology.create_connectivity(0, mesh.topology.dim)
v_to_c = mesh.topology.connectivity(0, mesh.topology.dim)
geom_nodes = dolfinx.mesh.entities_to_geometry(mesh, 0, np.arange(num_vertices,dtype=np.int32), False)
dof_layout = V.dofmap.dof_layout
vertex_to_dof_map = np.empty(num_vertices, dtype=np.int32)
num_cells = mesh.topology.index_map(
mesh.topology.dim).size_local + mesh.topology.index_map(
mesh.topology.dim).num_ghosts
c_to_v = mesh.topology.connectivity(mesh.topology.dim, 0)
for cell in range(num_cells):
vertices = c_to_v.links(cell)
dofs = V.dofmap.cell_dofs(cell)
for i, vertex in enumerate(vertices):
vertex_to_dof_map[vertex] = dofs[dof_layout.entity_dofs(0, i)]
u = dolfinx.fem.Function(V)
for i, node in enumerate(geom_nodes):
cells = v_to_c.links(i)
value = 0
for cell in cells:
value+=q.x.array[cell]
value/=len(cells)
u.x.array[vertex_to_dof_map[i]] = value
u.name = "P1"
u.x.scatter_forward()
with dolfinx.io.XDMFFile(mesh.comm, "output.xdmf", "w") as xdmf:
xdmf.write_mesh(mesh)
xdmf.write_function(u)
xdmf.write_function(q)
yielding