Hello everyone,
I am currently trying to solve a nonlinear problem by using petsc4py.PETSc.SNES directly. The problem is a 2D unit square with a neo-Hooke material and a traction applied to the right boundary. I followed the code given by niewiarowski and nate:
https://fenicsproject.discourse.group/t/using-petsc4py-petsc-snes-directly/2368
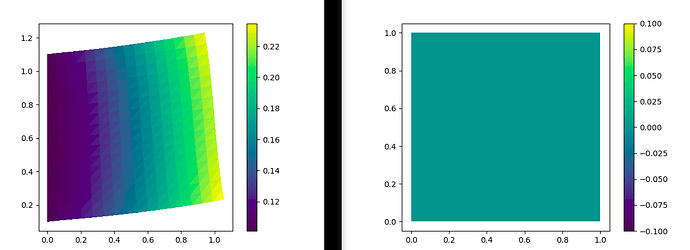
I encounter a problem when it comes to the boundary conditions: Everything seems to work fine when I set a non-homogeneous Dirichlet-BC (e.g. a displacement of (0,0.1) ) to the left boundary. However, when I set a zero-displacement Dirichlet-BC to the left boundary, the solution shows a zero-displacement in the whole domain, ignoring the traction set to the right side.
Here is a picture of the solution field, on the left with the non-zero Dirichlet-BC and on the right with the zero Dirichlet-BC:
Here is the MWE:
import dolfin as dlf
import numpy as np
import petsc4py, sys
petsc4py.init(sys.argv)
from petsc4py import PETSc
import matplotlib.pyplot as plt
# Elasticity parameters
E, nu = 10.0, 0.3
mu, lmbda = dlf.Constant(E/(2*(1 + nu))), dlf.Constant(E*nu/((1 + nu)*(1 - 2*nu)))
# boundary displacement
u0 = dlf.Constant((0, 0))
# u0 = dlf.Constant((0, 0.1)) # with this u0, everything seems to work
# boundary traction
t0 = dlf.Constant((0.0, 0.1))
# volume force
f = dlf.Constant((0.0, 0.0))
# define boundaries
def left(x, on_boundary):
return on_boundary and dlf.near(x[0], 0.0)
def right(x, on_boundary):
return on_boundary and dlf.near(x[0], 1)
mesh=dlf.UnitSquareMesh(16,16)
# define function space
V = dlf.VectorFunctionSpace(mesh, 'P', 1)
# define Dirichlet (displacement) boundary conditions
bcleft = dlf.DirichletBC(V, u0, left)
bcs = [bcleft]
#define Neumann (traction) boundary conditions
traction_markers = dlf.MeshFunction('size_t', mesh, mesh.topology().dim()-1)
traction_markers.set_all(0)
traction_domain = dlf.AutoSubDomain(right)
traction_domain.mark(traction_markers, 1)
ds_t = dlf.ds(subdomain_data=traction_markers)(1)
# define variational problem
u = dlf.Function(V)
v = dlf.TestFunction(V)
du = dlf.TrialFunction(V)
# Kinematics
d = u.geometric_dimension()
I = dlf.Identity(d) # Identity tensor
F = I + dlf.grad(u) # Deformation gradient
C = F.T*F
# Invariants of deformation tensors
Ic = dlf.tr(C)
J = dlf.det(F)
# Stored strain energy density (compressible neo-Hookean model)
psi = (mu/2)*(Ic - 3) - mu*dlf.ln(J) + (lmbda/2)*(dlf.ln(J))**2
# Total potential energy
Pi = psi*dlf.dx - dlf.dot(f, u)*dlf.dx - dlf.dot(t0, u)*ds_t
# Compute first variation of Pi (directional derivative about u in the direction of v)
Var = dlf.derivative(Pi, u, v)
#Jac = dlf.derivative(Var, u, du)
class SNESProblem():
def __init__(self, F, u, bcs):
V = u.function_space()
du = dlf.TrialFunction(V)
self.L = F
self.a = dlf.derivative(F, u, du)
self.bcs = bcs
self.u = u
def F(self, snes, x, F):
x = dlf.PETScVector(x)
F = dlf.PETScVector(F)
dlf.assemble(self.L, tensor=F)
for bc in self.bcs:
bc.apply(F, x)
def J(self, snes, x, J, P):
J = dlf.PETScMatrix(J)
dlf.assemble(self.a, tensor=J)
for bc in self.bcs:
bc.apply(J)
problem = SNESProblem(Var, u, bcs)
b = dlf.PETScVector()
J_mat = dlf.PETScMatrix()
snes = PETSc.SNES().create(dlf.MPI.comm_world)
snes.setFunction(problem.F, b.vec())
snes.setJacobian(problem.J, J_mat.mat())
snes.setFromOptions()
snes.solve(None, problem.u.vector().vec())
uconv = u((1,1))
print('')
print(' Reporting results ...')
print(' v(1/1) = ', uconv[1])
# Plot and hold solution
pu=dlf.plot(u, mode='displacement')
plt.colorbar(pu)
plt.show()
I am using the dolfin version 2019.2.0.dev.
Any help would be greatly appreciated!
Jana