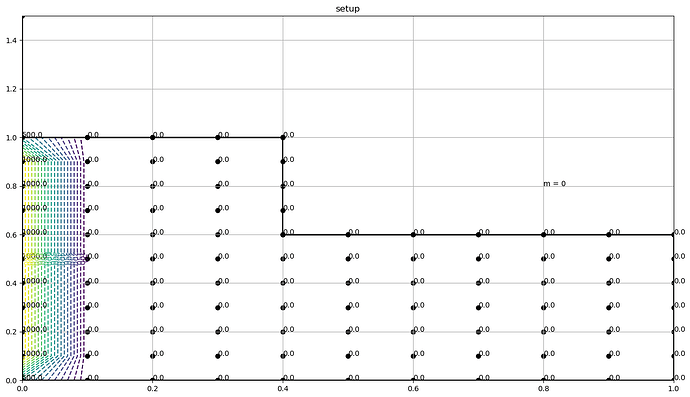
I have 2 questions which arise from wanting to apply Dirichlet boundary conditions to a L-shaped polygon like the one below
where it can be seen that the boundary values are 0 everywhere except for the left side of the L-shaped grid, where the corner values are 500 and the remaining values are 1000 (ignore the values on the points inside).
- Creating individual boundaries polygonal mesh?
I’ll start off creating the L-shaped mesh (which is the code of another user who had helped me previously) below:
bounds = [[0.0, 0.0], [1.0, 1.0]] # Original bounds of rectangle
xcutoff, ycutoff = 0.4, 0.6 # Cutoff to make L-shape
mesh = df.mesh.create_rectangle(MPI.COMM_WORLD, bounds, [10, 10])
tdim = mesh.topology.dim
im_tdim = mesh.topology.index_map(tdim)
cells = np.arange(im_tdim.size_local + im_tdim.num_ghosts, dtype=np.int32)
midpoints = df.mesh.compute_midpoints(mesh, tdim, cells)
cells_to_remove = (midpoints[:, 0] >= xcutoff) & (midpoints[:, 1] >= ycutoff)
cells_to_keep = np.where(~cells_to_remove)[0]
lmesh, _, _, _ = df.mesh.create_submesh(mesh, mesh.topology.dim, cells_to_keep)
And following the tutorial to create boundaries and attempting to apply this for an L-shaped block:
# Define the boundaries
boundaries = [(1, lambda x: np.isclose(x[0], 0)), #(marker = 1, locator = (x=0,y=y))
(2, lambda x: np.isclose(X[0],0.4) & np.greater_equal(X[1],0.6)), #(marker = 2, locator = (x=0.4,y>=0.6))
(3, lambda x: np.isclose(x[0], 1)), #(marker = 3, locator = (x=1,y=y))
(4, lambda x: np.isclose(x[1], 0)), #(marker = 4, locator = (x=x,y=0))
(5, lambda x: np.isclose(X[1],0.6) & np.greater_equal(X[0],0.4)), #(marker = 5, locator = (x>=0.4,y=0.6))
(6, lambda x: np.isclose(x[1], 1))] #(marker = 6, locator = (x=x,y=1))
My definitions for boundaries 2 and 5 raise issues at the step for identifying the facets which make up the boundaries in the next step
domain = lmesh
facet_indices, facet_markers = [], []
fdim = mesh.topology.dim - 1 # Facet dimensionality
for (marker, locator) in boundaries:
facets = df.mesh.locate_entities(domain, fdim, locator)
facet_indices.append(facets)
facet_markers.append(np.full_like(facets, marker))
gives the output error when reading in boundary 2:
---------------------------------------------------------------------------
RuntimeError Traceback (most recent call last)
Cell In[11], line 4
2 fdim = mesh.topology.dim - 1 # Facet dimensionality
3 for (marker, locator) in boundaries:
----> 4 facets = df.mesh.locate_entities(mesh, fdim, locator)
5 facet_indices.append(facets)
6 facet_markers.append(np.full_like(facets, marker))
File ~/anaconda3/envs/fea/lib/python3.13/site-packages/dolfinx/mesh.py:470, in locate_entities(msh, dim, marker)
456 def locate_entities(msh: Mesh, dim: int, marker: typing.Callable) -> np.ndarray:
457 """Compute mesh entities satisfying a geometric marking function.
458
459 Args:
(...) 468 Indices (local to the process) of marked mesh entities.
469 """
--> 470 return _cpp.mesh.locate_entities(msh._cpp_object, dim, marker)
RuntimeError: Length of array of markers is wrong.
Can my definitions for the boundaries be fixed, or will this require a different approach?
- Applying an array of values to a boundary?
I want to assign the array of values
vals = np.array([500,1000,1000,1000,1000,1000,1000,1000,1000,1000,500],dtype = 'int32')
to the left boundary to make it the same as in the figure. Previously I have seen how to apply an expression or a constant default_scalar_type(), but not an array of values to explicitly apply to each boundary point. Is there a way to do this?