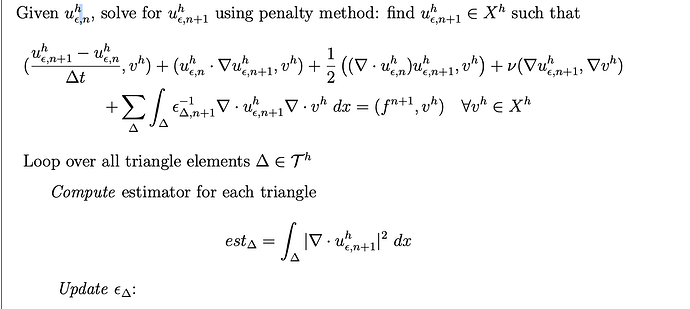Thank you so much for your help @francesco-ballarin. My bad, I should have uploaded some mini-code.
I am working on a time-dependent 2D Navier Stokes equation with an adaptive scheme, in which the penalty parameter varies for each cell, or we call it an element over the domain. My approach is to define a DG0 array that stores the penalty parameter for each cell. Then I solve the matrix, update my solution, and calculate some quantity on each cell as a criterion to update my penalty parameter for the next time step. Repeat this.
Do you think it will be reseaonale, I don’t know how to assemble the matrix while some coefficient is defined for each cell. I provide my code, which runs with no error if that helps.
Thank you!
from mpi4py import MPI
from petsc4py import PETSc
import dolfinx
from dolfinx import fem, mesh, io
from dolfinx.fem import Constant
from dolfinx.fem.assemble import assemble_matrix, create_vector, assemble_vector, apply_lifting, set_bc, assemble_scalar
import ufl
from ufl import div, dot, dx, inner, lhs, nabla_grad, rhs, grad
class Velocity:
def __init__(self):
self.t = 0.0
def eval(self, x):
# Added some spatial variation here.
return (np.pi * np.sin(self.t) * np.sin(2 * np.pi * x[1]) * (np.sin(np.pi * x[0])) ** 2,
-np.pi * np.sin(self.t) * np.sin(2 * np.pi * x[0]) * (np.sin(np.pi * x[1])) ** 2)
class Pressure:
def __init__(self):
self.t = 0.0
def eval(self, x):
return np.sin(self.t) *np.cos(np.pi * x[0]) * np.sin(np.pi * x[1])
class Source:
def __init__(self):
self.t=0.0
def eval(self, x):
return (np.pi*(4*np.pi**2*np.sin(self.t)**2*np.sin(np.pi*x[0])**3*np.sin(np.pi*x[1])*np.cos(np.pi*x[0]) +16*np.pi**2*np.sin(self.t)*np.sin(np.pi*x[0])**2*np.cos(np.pi*x[1]) - np.sin(self.t)*np.sin(np.pi*x[0]) - 4*np.pi**2*np.sin(self.t)*np.cos(np.pi*x[1]) + 2*np.sin(np.pi*x[0])**2*np.cos(self.t)*np.cos(np.pi*x[1]))*np.sin(np.pi*x[1]),
np.pi*(4*np.pi**2*np.sin(self.t)**2*np.sin(np.pi*x[0])**2*np.sin(np.pi*x[1])**3*np.cos(np.pi*x[1]) - 16*np.pi**2*np.sin(self.t)*np.sin(np.pi*x[0])*np.sin(np.pi*x[1])**2*np.cos(np.pi*x[0]) + 4*np.pi**2*np.sin(self.t)*np.sin(np.pi*x[0])*np.cos(np.pi*x[0]) + np.sin(self.t)*np.cos(np.pi*x[0])*np.cos(np.pi*x[1]) - 2*np.sin(np.pi*x[0])*np.sin(np.pi*x[1])**2*np.cos(self.t)*np.cos(np.pi*x[0])))
#exact solution of the test problem
u_exact = Velocity()
p_exact = Pressure()
f = Source()
# define mesh
nx = 2
ny = 2
msh = mesh.create_rectangle(MPI.COMM_WORLD, [np.array([-1, -1]), np.array([1, 1])], [nx, ny], dolfinx.cpp.mesh.CellType.triangle)
cell=msh.ufl_cell()
tdim = msh.topology.dim
num_cells = msh.topology.index_map(tdim).size_local
h = dolfinx.cpp.mesh.h(msh, tdim, range(num_cells)) #h is the largest edge
print("edge length", h)
DG0 = dolfinx.fem.FunctionSpace(msh, ("DG", 0))
v_DG0 = ufl.TestFunction(DG0)
# define function space
P2 = ufl.VectorElement("CG", cell, 2)
V = fem.FunctionSpace(msh, P2)
DG0_ufl = ufl.FiniteElement("DG", cell, 0)
V0 = fem.FunctionSpace(msh, DG0_ufl)
piecewise_coef =fem.Function(V0)
piecewise_coef.name = "piecewise_coef"
for i in range(len(piecewise_coef.x.array[:])):
piecewise_coef.x.array[i] = (i+1)*1e-3
"""useful expressions"""
def b(u, v, w): # Skew-symmetry nonlinear term
return .5 * (inner(dot(u, nabla_grad(v)), w) - inner(dot(u, nabla_grad(w)), v))
def a(u, v): # Viscous term (grad(u),grad(v))
return inner(nabla_grad(u), nabla_grad(v))
# define the velocity trial and test functions
u = ufl.TrialFunction(V)
v = ufl.TestFunction(V)
# define boundary conditions
from petsc4py.PETSc import ScalarType
uD = fem.Constant(msh, ScalarType((0.0,0.0)))
# Create facet to cell connectivity required to determine boundary facets
tdim = msh.topology.dim
fdim = tdim - 1
msh.topology.create_connectivity(fdim, tdim)
boundary_facets = mesh.exterior_facet_indices(msh.topology)
boundary_dofs = fem.locate_dofs_topological(V, fdim, boundary_facets)
bcu = fem.dirichletbc(uD, boundary_dofs,V)
bcs = [bcu]
# define the solution fields involved
u_exact_fem = fem.Function(V)
u_exact_fem.name = "u_exact_fem"
u_n = fem.Function(V)
u_n.name = "u_n"
u_next = fem.Function(V)
u_next.name = "u_next"
f_fem = fem.Function(V)
f_fem.name = "f_fem"
time_step = 0.1
nu = 1
t =0
t += time_step
#u_n
t += time_step
u_exact.t = t
u_n=fem.Function(V)
u_n.name = "u_n"
u_n.interpolate(u_exact.eval)
u_n.x.scatter_forward()
"""" algorithm """
F3 = 1.0 / time_step * inner(u, v) * dx
F3 += b(u_n, u, v) * dx
F3 += 1/piecewise_coef * div(u) * div(v) * dx #here
F3 += nu * a(u, v) * dx
F3 -= 1.0 / time_step * inner(u_n, v) * dx + inner(f_fem, v) * dx
a3 = fem.form(lhs(F3))
L3 = fem.form(rhs(F3))
A3 = fem.petsc.create_matrix(a3)
b3= fem.petsc.create_vector(L3)
#Direct Solver for NSE before turning on k equation
ksp3 = PETSc.KSP().create(msh.comm)
ksp3.setOperators(A3)
ksp3.setType("preonly")
pc3 = ksp3.getPC()
pc3.setType("lu")
pc3.setFactorSolverType("mumps")
pc3.setFactorSetUpSolverType()
# update time
t += time_step
f.t = t
u_exact.t = t
f_fem.interpolate(f.eval)
u_exact_fem.interpolate(u_exact.eval)
A3.zeroEntries()
fem.petsc.assemble_matrix(A3, a3, bcs=bcs)
A3.assemble()
with b3.localForm() as loc:
loc.set(0)
fem.petsc.assemble_vector(b3, L3)
fem.petsc.apply_lifting(b3, [a3], [bcs])
b3.ghostUpdate(addv=PETSc.InsertMode.ADD_VALUES, mode=PETSc.ScatterMode.REVERSE)
fem.petsc.set_bc(b3, bcs)
ksp3.solve(b3, u_next.vector)
u_next.x.scatter_forward()
divu_form = dolfinx.fem.form(inner(div(u_next), div(u_next))*v_DG0*ufl.dx)
divu_norm_element =dolfinx.fem.assemble_vector(divu_form )
print("francesco")
print(divu_norm_element.array)