Hi everyone, I have a working code that I want to improve, I simplified a lot to create a MWE.
I’m triying to couple Fenics-X via preCICE by myself, because the current adapter can solve 3D linear elastic problems at the moment.
So, to work with preCICE, I have to provide the mesh node coordinates of the coupling interface, then it will interpolate de compute data from the coupled solver, to that nodes. And I need to apply that computed data to the fenicsx model.
So in my code:
- I get the interface node coordinates directly from the gmsh model. but I want to get it from the mesh domain.
- Then I have in return (this is simulated in the MWE) an array of loads, in the same order of the nodes I give.
- I search the index of the nodes in the fenicsx mesh, by searching for the coords returned in step 1.
- finally I create a load Function over the FunctioSpace and map the array of loads to the function space.
At this point the code works but have a lot of issues. First the performance issue that implies extracting and the remapping the coordinates, as described in point 1, and 3. The other big issue is that this approach don’t work with a second order elelemt and function space.
import os
from typing import NamedTuple
import gmsh
import numpy as np
import pyvista
import ufl
from dolfinx import fem, io, mesh, plot
from dolfinx.fem.petsc import LinearProblem
from mpi4py import MPI
from petsc4py import PETSc
comm = MPI.COMM_WORLD
CURRENT_FOLDER = os.path.dirname(__file__)
# -------------------#
# Problem properties #
# -------------------#
class Material(NamedTuple):
name: str
E: PETSc.ScalarType
nu: PETSc.ScalarType
rho: PETSc.ScalarType
material = Material("Aluminum 6061", 68.9e9, 0.33, 2700)
# beam size
height = 150 # m
diameter = 6.5 # m
# ------- #
# MESHING #
# ------- #
FIXED_TAG = 1000
LOAD_SURFACE_TAG = 3000
VOLUME_TAG = 4000
ELEMENTS_ORDER = 1
def gmsh_tower(h: float, d: float) -> gmsh.model:
model_name = "Tower"
gmsh.model.add(model_name)
model = gmsh.model()
model.setCurrent(model_name)
# Recombine tetrahedra to hexahedra
gmsh.option.setNumber("Mesh.RecombinationAlgorithm", 2)
gmsh.option.setNumber("Mesh.RecombineAll", 2)
gmsh.option.setNumber("Mesh.CharacteristicLengthFactor", 0.1)
circle = model.occ.addDisk(0, 0, 0, d / 2, d / 2)
model.occ.extrude([(2, circle)], 0, 0, h, numElements=[h], recombine=True)
model.occ.synchronize()
fixed_sf = model.addPhysicalGroup(2, [1], tag=FIXED_TAG)
tip_sf = model.addPhysicalGroup(2, [2, 3], tag=LOAD_SURFACE_TAG)
vol = model.addPhysicalGroup(3, [1], tag=VOLUME_TAG)
model.setPhysicalName(2, fixed_sf, "FIXED SURFACE")
model.setPhysicalName(2, tip_sf, "LOAD SURFACE")
model.setPhysicalName(3, vol, "Mesh volume")
model.mesh.setOrder(ELEMENTS_ORDER)
model.mesh.generate(3)
# get node coordinates of LOAD_SURFACE physical group
boundaries = ((2, 2), (2, 3))
interface_nodes = []
for dim, tag in boundaries:
tag = abs(tag)
_, coords, _ = gmsh.model.mesh.getNodes(dim, tag, includeBoundary=True)
interface_nodes.extend(coords)
interface_nodes = np.array(interface_nodes).reshape((len(interface_nodes) // 3, 3))
return model, interface_nodes
# Create model
gmsh.initialize()
gmsh.option.setNumber("General.Terminal", 0)
model, interface_nodes = gmsh_tower(height, diameter)
domain, cell_markers, facet_markers = io.gmshio.model_to_mesh(model, comm, rank=0)
dim = domain.geometry.dim
domain.topology.create_connectivity(dim - 1, dim)
# -------------- #
# Function Space #
# -------------- #
V = fem.functionspace(domain, ("Lagrange", ELEMENTS_ORDER, (dim,)))
u = fem.Function(V, name="Displacement")
f = fem.Function(V, name="Force")
E = fem.Constant(domain, float(material.E))
nu = fem.Constant(domain, float(material.nu))
rho = fem.Constant(domain, float(material.rho))
# ------------------------------------ #
# APPLY DISTRIBUTED LOADS ON BEAM FACE #
# ------------------------------------ #
def map_mesh_ids(global_matrix, loads_matrix):
ids = []
for c in loads_matrix:
id = np.argwhere(np.all((global_matrix - c) == 0, axis=1)).item()
ids.append(id)
return ids
_loads_coords = interface_nodes
_domain_coords = domain.geometry.x
ids = map_mesh_ids(_domain_coords, _loads_coords)
# simulated preCICE response
external_loads = np.random.random(_loads_coords.shape) * 1000
_f = np.zeros(domain.geometry.x.shape)
_f[ids] = external_loads
f.x.array[:] = _f.flatten()
f.x.scatter_forward()
# --------------------#
# Boundary conditions #
# --------------------#
# Clamped
u_D = np.array([0, 0, 0], dtype=PETSc.ScalarType)
fixed_facets = facet_markers.find(FIXED_TAG)
fixed_surface_dofs = fem.locate_dofs_topological(V, 2, fixed_facets)
fixed_bc = fem.dirichletbc(u_D, fixed_surface_dofs, V)
# -------------------------#
# linear elastic equations #
# -------------------------#
load_facets = facet_markers.find(LOAD_SURFACE_TAG)
load_tags = mesh.meshtags(domain, dim - 1, np.sort(load_facets), 1)
ds = ufl.Measure("ds", domain=domain, subdomain_data=load_tags, subdomain_id=1)
lmbda = E * nu / (1 + nu) / (1 - 2 * nu)
mu = E / 2 / (1 + nu)
def epsilon(u):
return ufl.sym(ufl.grad(u))
def sigma(u):
return lmbda * ufl.nabla_div(u) * ufl.Identity(len(u)) + 2 * mu * epsilon(u)
du = ufl.TrialFunction(V)
u_ = ufl.TestFunction(V)
a = ufl.inner(sigma(du), epsilon(u_)) * ufl.dx
l = ufl.inner(f, u_) * ds(1)
bcs = [fixed_bc]
problem = LinearProblem(a, l, bcs=bcs, u=u, petsc_options={"ksp_type": "cg", "pc_type": "gamg"})
uh = problem.solve()
# ------------ #
# PLOT RESULTS #
# ------------ #
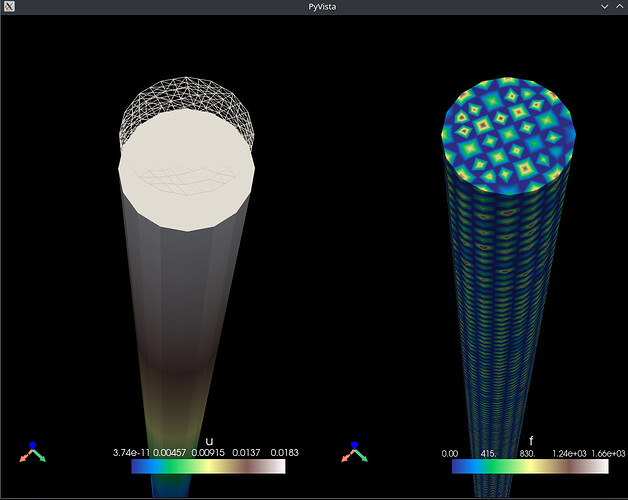
p = pyvista.Plotter(shape=(1, 2), theme=pyvista.themes.DarkTheme())
topology, cell_types, geometry = plot.vtk_mesh(V)
grid = pyvista.UnstructuredGrid(topology, cell_types, geometry)
grid["u"] = u.x.array.reshape((geometry.shape[0], 3))
grid["f"] = f.x.array.reshape((geometry.shape[0], 3))
p.subplot(0, 0)
grid.set_active_scalars("u")
p.add_mesh(grid.copy(), style="wireframe", cmap="terrain")
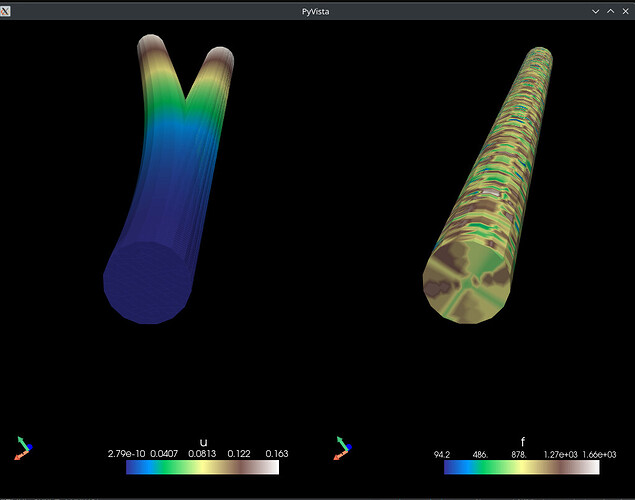
warped = grid.warp_by_vector("u", factor=100)
p.add_mesh(warped, show_edges=False, cmap="terrain")
p.add_axes(line_width=5, cone_radius=0.6, shaft_length=0.7, tip_length=0.3)
p.subplot(0, 1)
grid.set_active_scalars("f")
p.add_mesh(grid.copy(), show_edges=False, cmap="terrain")
p.add_axes(line_width=5, cone_radius=0.6, shaft_length=0.7, tip_length=0.3)
p.link_views()
p.show()
Result: