Hello.
I used docker to run with dolfinx and now implementing code in jupyter lab environment.
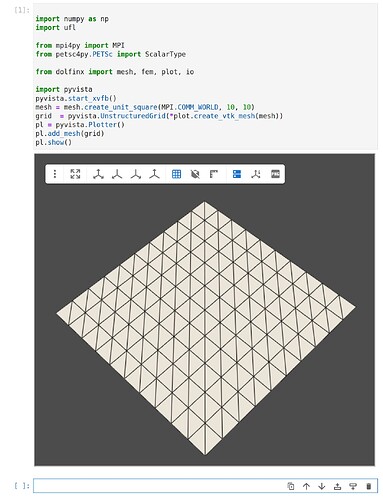
However, I found error in displaying result visualization. My code is as follows (Linerar elasiticity demo code)
import numpy as np
import ufl
from mpi4py import MPI
from petsc4py.PETSc import ScalarType
from dolfinx import mesh, fem, plot, io
import ipyvtklink
import pyvista
pyvista.start_xvfb()
For visualization part,
# Create plotter and pyvista grid
p = pyvista.Plotter(notebook=True)
topology, cell_types, geometry = plot.create_vtk_mesh(V)
grid = pyvista.UnstructuredGrid(topology, cell_types, geometry)
# Attach vector values to grid and warp grid by vector
grid["u"] = uh.x.array.reshape((geometry.shape[0], 3))
actor_0 = p.add_mesh(grid, style="wireframe", color="k")
warped = grid.warp_by_vector("u", factor=1.5)
actor_1 = p.add_mesh(warped, show_edges=True)
p.show_axes()
if not pyvista.OFF_SCREEN:
p.show()
else:
figure_as_array = p.screenshot("deflection.png")
However, the result is
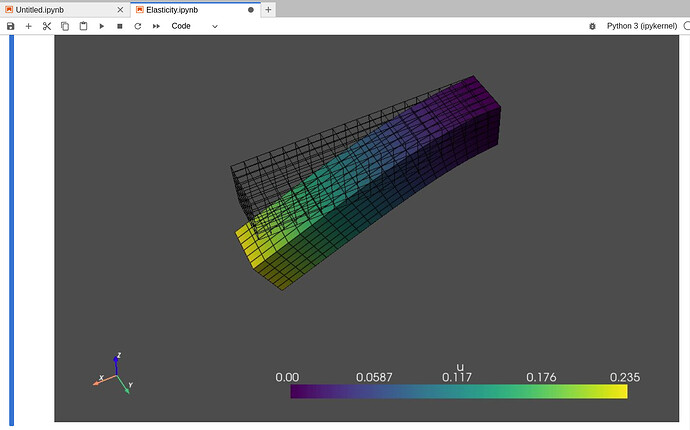
Actually, I could get visualization, but I could not interact with the result. So, I installed jupyter backend ‘trame’ for pyvista, and the result is like this.
Could you help me to solve this issue?
Thanks for reading.