I’m sorry, I misread your post and hastily typed out a reply earlier. So, we want to use a separate function space to interpolate our sources to to create a function, and a different function space for the test and trial functions. This is similar to what was done in an older example I found here where f and g and the test and trial function each have their own respective function spaces.
Also, to your confusion, my true problem is one such that I have a dataset in which I don’t know what the symbolic or analytic definition of the data is, and I needed to know how to handle this. I think you answered that as well by saying that I need to interpolate this data which is exactly what Nate’s code did.
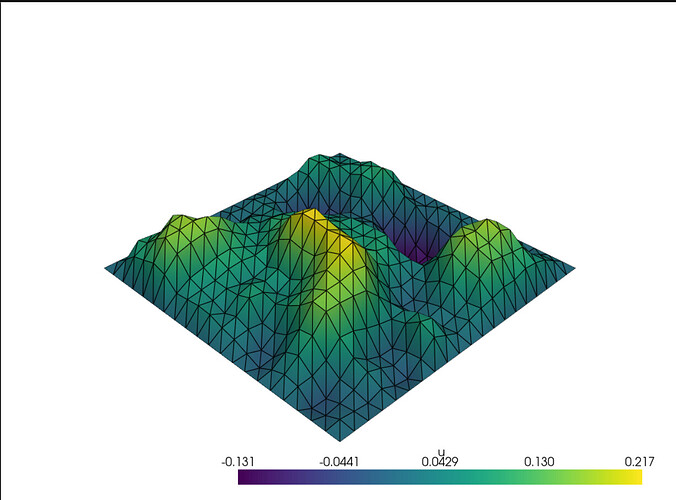
Making the necessary changes to my code, this did work… I think? Let me know what you think.
# Create mesh
domain = df.mesh.create_unit_square(MPI.COMM_WORLD,20,20)
tdim = domain.topology.dim
fdim = domain.topology.dim-1
domain.topology.create_connectivity(fdim, tdim)
# Create functionspace for Test and Trial functions
V = df.fem.functionspace(mesh = domain, element = ("Lagrange",1))
# Define Trial and Test functions
u = ufl.TrialFunction(V)
s = ufl.TestFunction(V)
# Create separate functionspace for forcing term
DG0 = df.fem.functionspace(domain, ("DG", 0))
# Define forcing function
f = df.fem.Function(DG0)
f_size_local = DG0.dofmap.index_map.size_local
f.x.array[:f_size_local] = np.random.uniform(-100, 100, size=f_size_local)
f.x.scatter_forward()
# Define boundary
boundary_facets = df.mesh.exterior_facet_indices(domain.topology)
boundary_dofs = df.fem.locate_dofs_topological(V = V, entity_dim=1, entities= boundary_facets)
# Define BC given boundary_dofs
uD = df.fem.dirichletbc(df.default_scalar_type(0),boundary_dofs,V)
# Abstract form of variational problem
## a(u,s)
a = ufl.dot(ufl.grad(u),ufl.grad(s)) * ufl.dx
## L(s)
L = f*s*ufl.dx
# define problem
from dolfinx.fem.petsc import LinearProblem
problem = LinearProblem(a=a,L=L,bcs = [uD])
# solve
uh = problem.solve()
# Plot
u_topology, u_cell_types, u_geometry = df.plot.vtk_mesh(V.mesh)
u_grid = pv.UnstructuredGrid(u_topology, u_cell_types, u_geometry)
u_grid.point_data["u"] = uh.x.array.real
warped = u_grid.warp_by_scalar("u", factor=1) # 3D warping of solution
u_grid.set_active_scalars("u")
u_plotter = pv.Plotter()
u_plotter.add_mesh(warped, show_edges=True)
u_plotter.show()
Notice how I changed the line defining populating random values into f to
f.x.array[:f_size_local] = np.random.uniform(-100, 100, size=f_size_local) so that the low and high values of the randomizer to be equal and opposite sign. When the distribution of values are a bit more symmetric about 0, I notice I get a random distribution of relative maxima and minima, which I think is what I should expect for a random distribution of sources. However, when I played around with the bounds, I noticed that the more skewed the bounds were away from 0, I would get a larger, smoother, more centered Gaussian hump. Running the original line
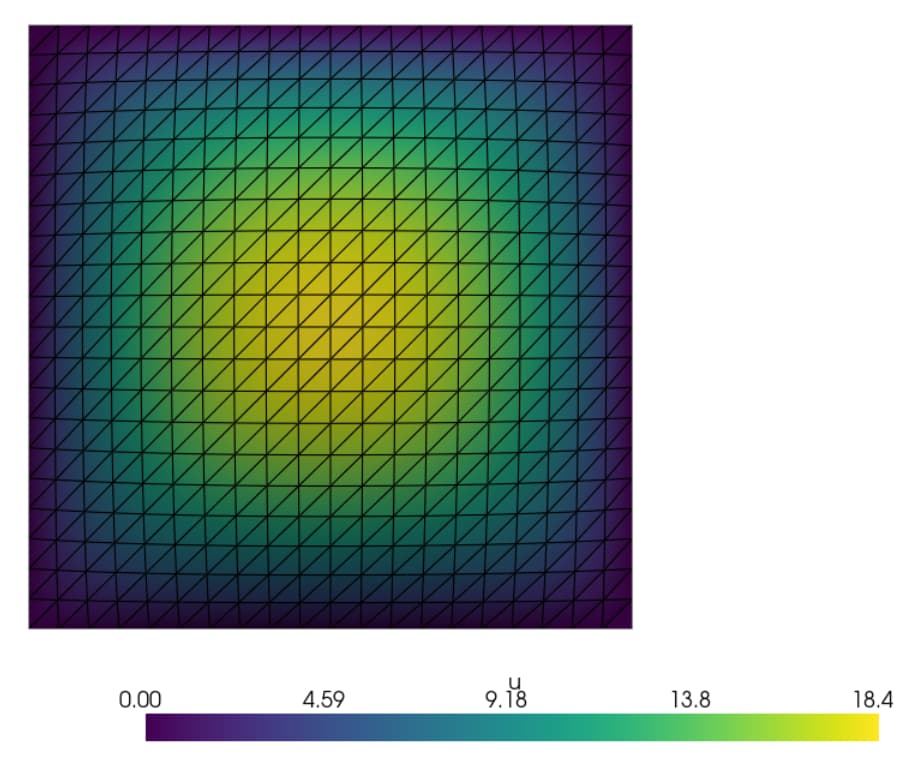
f.x.array[:f_size_local] = np.random.uniform(200, 300, size=f_size_local)
(where both bounds are positive), I get
I think that regardless of the bounds, I should expect a random bumpier distribution of multiple relative maxima and minima like the first plot given a random distribution of sources, not a singular smooth centered hump.