Hi,
I am working on error convergence analysis for an diffusion example in 2D : Cube is the domain and initial condition is a point source at the centre. Below is the formulation and analytical solution. I am approximating point source as Gaussian function.
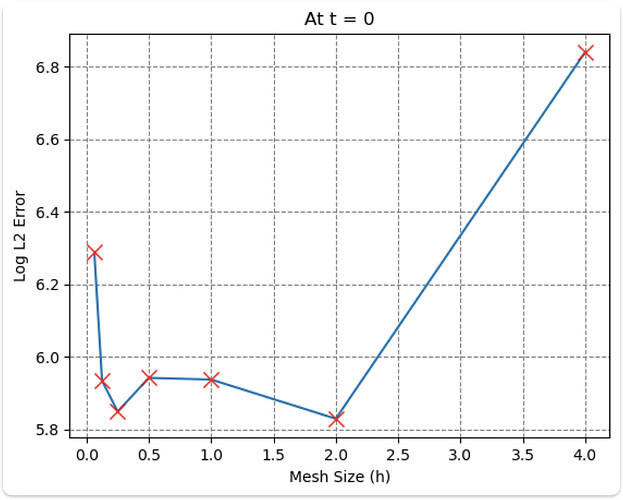
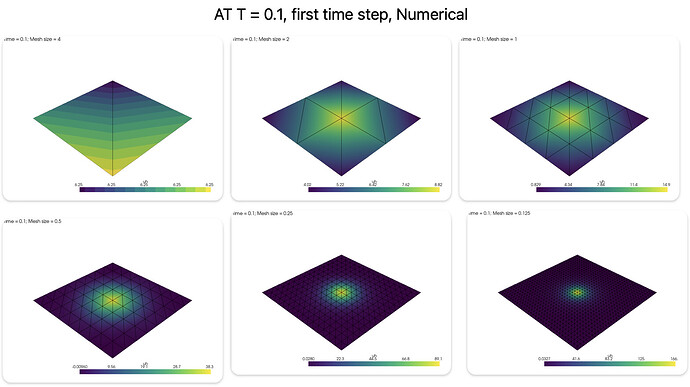
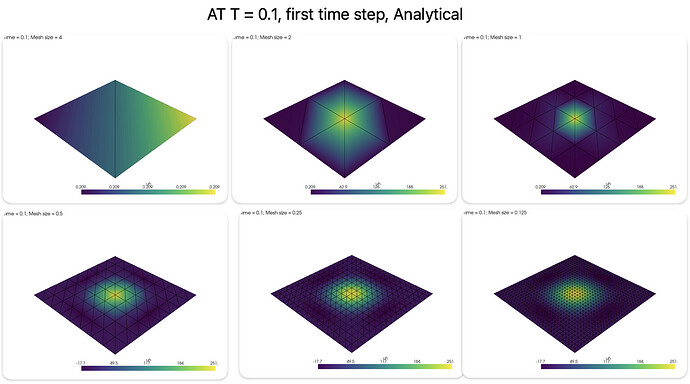
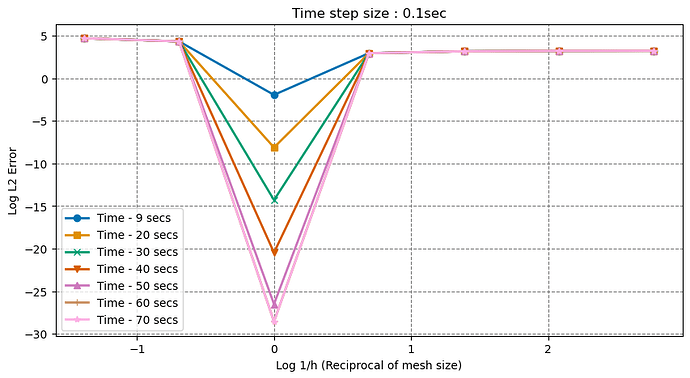
But the L2 error is increasing as I decrease the mesh size, can’t seem to find why this is happening. Tried changing the time w.r.t mesh size, but it didn’t work.
Here is the code:
import matplotlib as mpl
import pyvista
import ufl
import numpy as np
from petsc4py import PETSc
from mpi4py import MPI
from dolfinx import fem, mesh, io, plot
from dolfinx.fem.petsc import assemble_vector, assemble_matrix, create_vector, apply_lifting, set_bc
from dolfinx.fem import (Expression, Function, functionspace,
assemble_scalar, dirichletbc, form, locate_dofs_topological)
class exact_solution():
def __init__(self, t, cnt, dim, S0 =100):
self.S0 = S0
self.t = t
self.cnt = cnt
self.dim = dim
def __call__(self, x):
sol_approx = 0
for i in range(self.cnt):
for j in range(self.cnt):
norm_factor = (2 if i > 0 else 1) * (2 if j > 0 else 1) ## Normalization factor for the Fourier series terms
lambda2 = ((np.pi**2) * (i**2 + j**2))/16 ## Eigenvalue for the Fourier series terms
sol_approx += (self.S0/16) * norm_factor * np.cos(i*np.pi*x[0]/4) * np.cos(j*np.pi*x[1]/4) * np.exp(-lambda2 * self.t) ## Fourier series term
return sol_approx
class heat_eq_2D():
def __init__(self, domain, strt_t, end_t, num_steps,cnt,uexact,u_D,sigma):
self.domain = domain
self.V = fem.functionspace(domain, ("Lagrange", 1))
self.dt = end_t / num_steps
self.t = strt_t
self.cnt = cnt
self.uexact = uexact
self.u_D = u_D
self.num_steps = num_steps
self.sigma = sigma
def initial_condition(self, x,scale=1,S0=100):
return scale*S0*np.exp(-0.5*(x[0]**2 + x[1]**2)/(self.sigma**2) )/(2*np.pi*self.sigma**2)
def scale_est(self):
u_n0 = fem.Function(self.V)
u_n0.interpolate(lambda x: self.initial_condition(x,S0 = 1))
dx = ufl.dx(domain=self.domain)
inital_condt_form = u_n0*dx
integral = self.domain.comm.allreduce(fem.assemble_scalar(fem.form(inital_condt_form)), op=MPI.SUM)
self.scale = 1/integral
# print("Scale factor: ", self.scale)
def error_L2(self, uh, u_ex, degree_raise=3):
# Create higher order function space
degree = uh.function_space.ufl_element().degree
family = uh.function_space.ufl_element().family_name
mesh = uh.function_space.mesh
W = functionspace(mesh, (family, degree + degree_raise))
# Interpolate approximate solution
u_W = Function(W)
u_W.interpolate(uh)
# Interpolate exact solution, special handling if exact solution
# is a ufl expression or a python lambda function
u_ex_W = Function(W)
u_ex_W.interpolate(u_ex)
# Compute the error in the higher order function space
e_W = Function(W)
e_W.x.array[:] = u_W.x.array - u_ex_W.x.array
# Integrate the error
error = form(ufl.inner(e_W, e_W) * ufl.dx)
error_local = assemble_scalar(error)
error_global = mesh.comm.allreduce(error_local, op=MPI.SUM)
return np.sqrt(error_global)
def solve_pde(self):
self.scale_est()
u_n = fem.Function(self.V)
u_n.name = "u_n"
u_n.interpolate(lambda x: self.initial_condition(x, scale = self.scale))
dx = ufl.dx(domain=self.domain)
inital_condt_form = u_n*dx
integral = self.domain.comm.allreduce(fem.assemble_scalar(fem.form(inital_condt_form)), op=MPI.SUM)
# print(f"Integral of initial condition: {integral}")
# Define solution variable, and interpolate initial solution for visualization in Paraview
uh = fem.Function(self.V)
uh.name = "uh"
uh.interpolate(lambda x: self.initial_condition(x, scale=self.scale))
u, v = ufl.TrialFunction(self.V), ufl.TestFunction(self.V)
a = u * v * ufl.dx + self.dt * ufl.dot(ufl.grad(u), ufl.grad(v)) * ufl.dx
L = u_n * v * ufl.dx
bilinear_form = fem.form(a)
linear_form = fem.form(L)
A = assemble_matrix(bilinear_form)
A.assemble()
b = create_vector(linear_form)
solver = PETSc.KSP().create(self.domain.comm)
solver.setOperators(A)
solver.setType(PETSc.KSP.Type.PREONLY)
solver.getPC().setType(PETSc.PC.Type.LU)
time_ls = []
error_L2_ls = []
# error_max_ls = []
for i in range(self.num_steps):
self.t += self.dt
self.uexact.t += self.dt
self.u_D.interpolate(self.uexact)
# Update the right hand side reusing the initial vector
with b.localForm() as loc_b:
loc_b.set(0)
assemble_vector(b, linear_form)
# Solve linear problem
solver.solve(b, uh.x.petsc_vec)
uh.x.scatter_forward()
# Compute L2 error and error at nodes
# V_ex = fem.functionspace(self.domain, ("Lagrange", 1))
# u_ex = fem.Function(V_ex)
# u_ex.interpolate(self.uexact)
# error_L2 = np.sqrt(domain.comm.allreduce(fem.assemble_scalar(fem.form((uh - u_ex)**2 * ufl.dx)),
# op=MPI.SUM))
error_L2_ls.append(self.error_L2(uh,self.uexact))
time_ls.append(self.t)
# # Compute values at mesh vertices
# error_max = self.domain.comm.allreduce(np.max(np.abs(uh.x.array - self.u_D.x.array)), op=MPI.MAX)
# error_max_ls.append(error_max)
# Update solution at previous time step (u_n)
u_n.x.array[:] = uh.x.array
# self.end_num_sol = uh.x.array
# self.end_ana_sol = u_ex.x.array
# return error_L2_ls, error_max_ls
return error_L2_ls,time_ls
# Define temporal parameters
t = 0 # Start time
T = 2*60 # Final time
L2_error_dict_master = {}
for dt in [1,0.5,0.1,0.01,0.001]:
L2_error_dict = {}
for h in [1,0.8,0.5,0.25]:
print(f"Solving for mesh size = {h}")
num_steps = int(T/dt)
# Define mesh
nx, ny = int(4/h), int(4/h) # Number of elements in x and y directions
domain = mesh.create_rectangle(MPI.COMM_WORLD, [np.array([-2, -2]), np.array([2, 2])],
[nx, ny], mesh.CellType.triangle)
V = fem.functionspace(domain, ("Lagrange", 1))
u_exact = exact_solution(0,cnt = 5)
u_D = fem.Function(V)
u_D.interpolate(u_exact)
pde_sol_obj = heat_eq_2D(domain, strt_t=0, end_t=T, num_steps=num_steps,
cnt=5, uexact=u_exact, u_D=u_D, sigma=h)
L2_error_dict[h] = pde_sol_obj.solve_pde()
L2_error_dict_master[dt] = L2_error_dict