Dear all.
So, I played around with the model and found my personal best ‘convincing’ solution … so far.  A full example is shown here, which is a minimal MWE of the current code:
A full example is shown here, which is a minimal MWE of the current code:
import matplotlib.pyplot as plt
import numpy as np
import scipy.constants as const
import os
# _____________________________________________________ The values to play with
# Main parameters
pot_sphere = 3.0 # Potential of the sphere
pot_plane = 0.0 # Potential of the plane
sphere_r = 3.0 # Radius of the sphere
domain_size = 80.0 # Domain size (sphere)
plane_size = 70.0 # Plane size
plane_height = 0.5
scene_z_off = -16.0 # Z offset value for the sphere and plane
z = 1.0 # Distance sphere-plane (in nm)
mesh_prec_domain = 6.0 # Precision of the mesh in the domain
# I use a bounding mesh box with a refined mesh around the sphere
mesh_prec_ball = 0.3 # Mesh precision
# mesh_box_sl: lateral size in %, 1.0 = lateral size of the plane
# mesh_box_sh: height in %,
# first value : towrads the plane, 1.0 = full height
# second value: towrads the sphere, 1.0 = complete sphere is included
mesh_box_sl = 0.15
mesh_box_sh = (1.0, 1.0)
# Other things
sphere_x = 0.0
sphere_y = 0.0
sphere_z = 0.0
plane_x = 0.0
plane_y = 0.0
plane_z = 0.0
plane_vx = 0.0
plane_vy = 0.0
plane_vz = 0.0
# ___________________________________________________________________ Directory
text_spaces = " "
dir_base = "./" + os.path.basename(__file__)[:-3]
if not os.path.isdir(dir_base):
os.mkdir(dir_base)
# ____________________________________________________ Mesh creation via pygmsh
sphere_z = sphere_r + z
# The position and size of the plane
plane_z = -plane_height
plane_vz = plane_height
# Build the mesh first with pygmsh
geom = pygmsh.opencascade.Geometry()
# Create the domain, sphere and plane, and ...
# The domain is a large sphere.
domain = geom.add_ball([0.0, 0.0, 0.0],
domain_size)
sphere = geom.add_ball([sphere_x,
sphere_y,
sphere_z+scene_z_off],
sphere_r)
plane = geom.add_cylinder([plane_x,
plane_y,
plane_z+scene_z_off],
[plane_vx,
plane_vy,
plane_vz],
plane_size,
2.0*const.pi)
# ... take the difference (boolean).
space_1 = geom.boolean_difference([domain],[sphere],
delete_first=True,
delete_other=True)
space_2 = geom.boolean_difference([space_1],[plane],
delete_first=True,
delete_other=True)
# Sphere
geom.add_raw_code("Physical Surface(\"1\") = {7};")
# Plane below sphere
geom.add_raw_code("Physical Surface(\"2\") = {3,4,5};")
# Domain
geom.add_raw_code("Physical Surface(\"3\") = {6};")
# Volume
geom.add_raw_code("Physical Volume(\"4\") = {" + space_2.id + "};")
# The refined mesh in a bounding box around the sphere and the plane.
center_z = scene_z_off + z/2.0
z_min = center_z - z/2.0 - mesh_box_sh[0] * plane_height
z_max = center_z + z/2.0 + mesh_box_sh[1] * 2.0 * sphere_r
x_max = (mesh_box_sl * plane_size) / 2.0
x_min = -(mesh_box_sl * plane_size) / 2.0
y_max = x_max
y_min = x_min
# We hard-code the refined mesh into the .geo file via the Field option.
geom.add_raw_code("//+")
geom.add_raw_code("Field[1] = Box;")
geom.add_raw_code("//+")
geom.add_raw_code("Field[1].VIn = " + str(mesh_prec_ball) + ";")
geom.add_raw_code("//+")
geom.add_raw_code("Field[1].VOut = " + str(mesh_prec_domain) + ";")
geom.add_raw_code("//+")
geom.add_raw_code("Field[1].XMax = " + str(x_max) + ";")
geom.add_raw_code("//+")
geom.add_raw_code("Field[1].XMin = " + str(x_min) + ";")
geom.add_raw_code("//+")
geom.add_raw_code("Field[1].YMax = " + str(y_max) + ";")
geom.add_raw_code("//+")
geom.add_raw_code("Field[1].YMin = " + str(y_min) + ";")
geom.add_raw_code("//+")
geom.add_raw_code("Field[1].ZMax = " + str(z_max) + ";")
geom.add_raw_code("//+")
geom.add_raw_code("Field[1].ZMin = " + str(z_min) + ";")
geom.add_raw_code("//+")
geom.add_raw_code("Background Field = 1;")
file_geo = os.path.join(dir_base, "Geometry.geo")
mesh = pygmsh.generate_mesh(geom, geo_filename=file_geo)
# ______________________________________________ Mesh conversion and re-reading
# What follows is this mesh conversion stuff ... .
tetra_cells = None
tetra_data = None
triangle_cells = None
triangle_data = None
for key in mesh.cell_data_dict["gmsh:physical"].keys():
if key == "triangle":
triangle_data = mesh.cell_data_dict["gmsh:physical"][key]
elif key == "tetra":
tetra_data = mesh.cell_data_dict["gmsh:physical"][key]
for cell in mesh.cells:
if cell.type == "tetra":
if tetra_cells is None:
tetra_cells = cell.data
else:
tetra_cells = np.vstack([tetra_cells, cell.data])
elif cell.type == "triangle":
if triangle_cells is None:
triangle_cells = cell.data
else:
triangle_cells = np.vstack([triangle_cells, cell.data])
tetra_mesh = meshio.Mesh(points=mesh.points,
cells={"tetra": tetra_cells},
cell_data={"name_to_read":[tetra_data]})
triangle_mesh =meshio.Mesh(points=mesh.points,
cells=[("triangle", triangle_cells)],
cell_data={"name_to_read":[triangle_data]})
meshio.write(os.path.join(dir_base,"tetra_mesh.xdmf"), tetra_mesh)
meshio.write(os.path.join(dir_base,"mf.xdmf") , triangle_mesh)
# All the latter .xdmf files are read such that they can be used in FEniCS.
mesh = Mesh()
with XDMFFile(os.path.join(dir_base,"tetra_mesh.xdmf")) as infile:
infile.read(mesh)
mvc = MeshValueCollection("size_t", mesh, 2)
with XDMFFile(os.path.join(dir_base,"mf.xdmf")) as infile:
infile.read(mvc, "name_to_read")
boundaries = cpp.mesh.MeshFunctionSizet(mesh, mvc)
mvc2 = MeshValueCollection("size_t", mesh, 3)
with XDMFFile(os.path.join(dir_base,"tetra_mesh.xdmf")) as infile:
infile.read(mvc2, "name_to_read")
subdomains = cpp.mesh.MeshFunctionSizet(mesh, mvc2)
# ______________________________________ (Sub)domains, boundary conditions, etc.
dx = Measure("dx", domain=mesh, subdomain_data=subdomains)
ds = Measure("ds", domain=mesh, subdomain_data=boundaries)
# The mesh function.
V0 = FunctionSpace(mesh, "DG", 0)
V = FunctionSpace(mesh, "P", 2)
#V = FunctionSpace(mesh, "P", 2)
b_cond = []
#b_cond.append(DirichletBC(V, Constant(0.0), boundaries, 3)) # Main domain (large sphere)
b_cond.append(DirichletBC(V, Constant(pot_sphere), boundaries, 1)) # 1: sphere
b_cond.append(DirichletBC(V, Constant(pot_plane), boundaries, 2)) # 2: plane
# Domain boundary (large sphere): Robin condition
# From: Basaran, Scriven, Phys. Fluids A Fluid Dyn. 1, 795 (1989).
#r = Constant(2.0)
r = Expression("sqrt(" \
"pow(x[0]-rx, 2) + " \
"pow(x[1]-ry, 2) + " \
"pow(x[2]-rz, 2))", degree=1,
rx=sphere_x, ry=sphere_y, rz=sphere_z)
p = 1 / r
q = Constant(0.0)
# Poisson equation with the source set to 0
u = TrialFunction(V)
v = TestFunction(V)
a = dot(grad(u), grad(v)) * dx + p * u * v * ds(3)
L = Constant('0') * v * dx + p * q * v * ds(3)
u = Function(V)
solve(a == L, u, b_cond, solver_parameters = {"linear_solver": "gmres",
"preconditioner": "ilu"})
electric_field = project(-grad(u))
potentialFile = File(os.path.join(dir_base,"Potential.pvd"))
potentialFile << u
e_fieldfile = File(os.path.join(dir_base,"Efield.pvd"))
e_fieldfile << electric_field
# Force (in nN), calculated from the potential.
# ds(1) -> boundary No. 1 -> the sphere (see above)
# n[2] -> z-component of the normal vector
n = FacetNormal(mesh)
force_n = assemble( dot(grad(u), grad(u))*n[2]*ds(1) )
force_n = force_n * (const.epsilon_0/2.0)
print("\n\n" + text_spaces + "Force, numerical: " + str(force_n))
# Here is the analytical result for the force. The equation was taken from
# S. Hudlet, M. Saint Jean, C. Guthmann, J. Berger, Eur. Phys. J. B 2, 5 (1998)
# F = Pi * eps_0 * ( R^2/(z*(z+R)) ) * V^2
dist_z = sphere_z-sphere_r
dist_f = sphere_r**2.0 / (dist_z * (dist_z+sphere_r))
force_a = const.pi * const.epsilon_0 * dist_f * (pot_plane - pot_sphere)**2.0
print(text_spaces + "Force, analytical: " + str(force_a) + "\n")
print(text_spaces + "Error (Fa-Fn)/Fa: " + str((force_a-force_n)/force_a*100.0) + " %\n")
I’m not really a 100% sure if I have done correctly the coding, also in an ‘intelligent way’, may be you have a look. With respect to the previous code from above, I changed the following things:
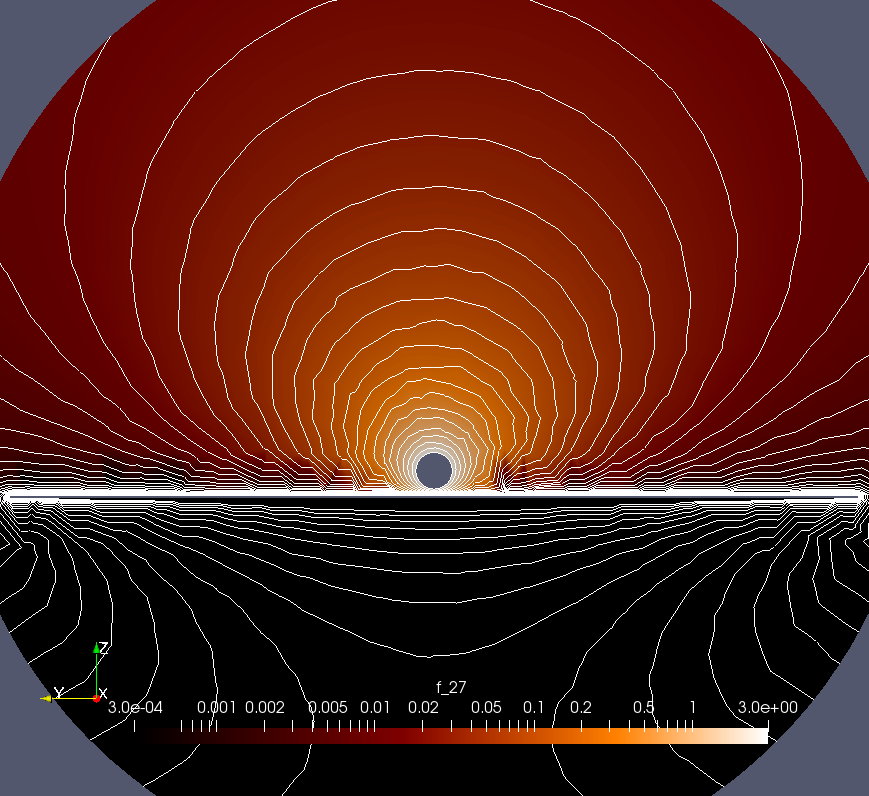
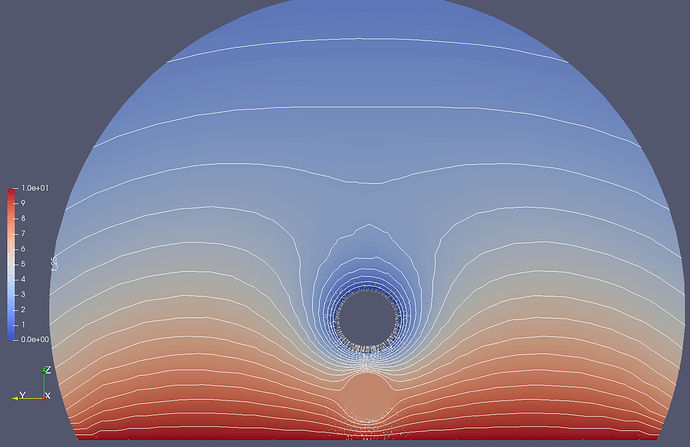
- I have chosen a spherical geometry: the domain is now a large sphere, and also the plane is a circular disk (thin cylinder) now (see image 1). This creates much better well-formed equipotential lines (see image 2, logarithmic color scale and scale for the lines!)
- The mesh of the region around the sphere and the plane underneath is now represented in a much finer mesh. This improves much the force value because most of the sphere-plane interaction is created there (see image 3). In fact, I hard-code via
geom.add_raw_code a box with properties mesh_box_sl (lateral size) and mesh_box_sh (height) into the .geo file with the help of Field[1] = Box; etc. In my code, the mesh’s precision is is given by mesh_prec_ball whereas mesh_prec_domain is the precision of the mesh in the rest of the domain.
-
I now use gmres as a linear solver and ilu as a pre-conditioner. With them, I can make use of fine meshes without ‘OUT OF MEMORY’ issues. Other linear solvers do not really improve the ‘OUT OF MEMORY’ issue but I’m still working on this.
-
I apply a Robin boundary condition on the domain boundary (large sphere) with dphi/dr = phi/r, as described in Basaran, Scriven, Phys. Fluids A Fluid Dyn. 1, 795 (1989).
All these changes produce a much smaller error which is now around 8% with respect to the expected analytical force value, in the distance range of interest (between 0.5 and 3 nm).
Force, numerical: 5.189529363356381e-10
Force, analytical: 5.632790908771443e-10
Error (Fa-Fn)/Fa: 7.869305866205872 %
Questions
Q1: Does the mesh, which I’m using, make sense? I have the following statistics:
Input points: 8521
Input facets: 17030
Input segments: 25545
Input holes: 0
Input regions: 0
Mesh points: 8521
Mesh tetrahedra: 41081
Mesh faces: 85247
Mesh edges: 52686
Mesh faces on facets: 17030
Mesh edges on segments: 25545
Is the mesh size still too large?
When decreasing the mesh size in the domain, not much changes. It is only in the region around the sphere that is quite sensitive to the mesh size, which makes sense because most of the sphere-plane interaction is created there … . To my opinion, one has to increase even more the mesh. What do you think?
Q2: The latter brings me to the limits of my laptop. With the parameters from above I’m at the full limit: if I lower the mesh size a bit I obtain an ‘OUT OF MEMORY’.
Is it possible to calculate only, e.g., a quarter of the sphere-plane structure since the electrostatic field and potential is highly symmetric around the z-axis?
Other (bad) idea: if I model the sphere-plane in 2D, I will not obtain the right force, right? In other words, a 3D case needs to be modeled in 3D in FEM, is this right?
Q3: I would like to play with the parameters of the solver and have read the manual Solving PDEs in Python – The FEniCS Tutorial Volume I (page 119). How can I change the following code
u = Function(V)
problem = LinearVariationalProblem(a, L, u, bc)
solver = LinearVariationalSolver(problem)
solver.parameters.linear_solver = ’gmres’
solver.parameters.preconditioner = ’ilu’
prm = solver.parameters.krylov_solver # short form
prm.absolute_tolerance = 1E-7
prm.relative_tolerance = 1E-4
prm.maximum_iterations = 1000
solver.solve()
such that it works? I obtain an error at prm = solver.parameters.krylov_solver
Because I’m still quite new to FEM applications I still need some help. Any help is highly appreciated. Thanks in advance. 





 A full example is shown here, which is a minimal MWE of the current code:
A full example is shown here, which is a minimal MWE of the current code: