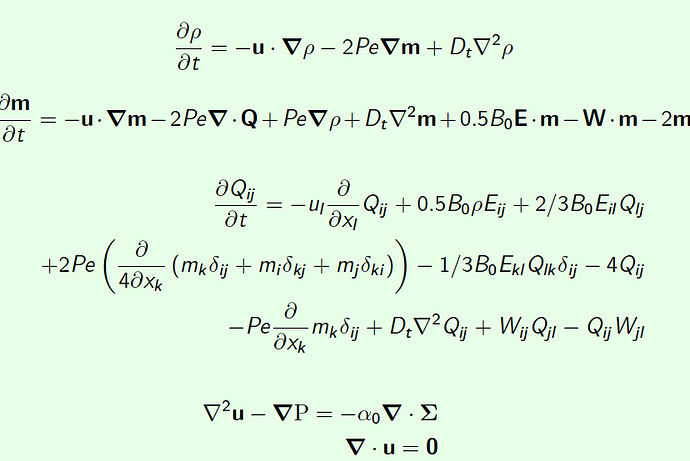Hi, thanks for reply.
This is my system of equations :
density (rho)
\begin{equation*}
\frac{\partial \rho}{\partial t} = -\mathbf{u}\cdot\boldsymbol{\nabla}\rho -2Pe\boldsymbol{\nabla}\mathbf{m} + D_{t}\nabla^{2}\rho
\end{equation*}
polarization vector m
\begin{equation*}
\frac{\partial \mathbf{m}}{\partial t} = -\mathbf{u}\cdot\boldsymbol{\nabla}\mathbf{m} -2Pe\boldsymbol{\nabla}\cdot\mathbf{Q} + Pe\boldsymbol{\nabla}\rho + D_{t}\nabla^{2}\mathbf{m} + 0.5B_{0}\mathbf{E}\cdot\mathbf{m} - \mathbf{W}\cdot\mathbf{m} - 2\mathbf{m}
\end{equation*}
neamtic order tensor Q,
\begin{align*}
\frac{\partial Q_{ij}}{\partial t} = - u_{l}\frac{\partial}{\partial x_{l}} Q_{ij} +0.5B_{0}\rho E_{ij} + 2/3B_{0} E_{il}Q_{lj}\ + 2Pe\left( \frac{\partial}{4\partial x_{k}}\left( m_{k}\delta_{ij} + m_{i}\delta_{kj} + m_{j}\delta_{ki} \right) \right) -1/3B_{0} E_{kl}Q_{lk}\delta_{ij}- 4 Q_{ij}\
-Pe\frac{\partial}{\partial x_{k}}m_{k}\delta_{ij}+D_{t}\nabla^{2}Q_{ij} + W_{ij}Q_{jl}-Q_{ij}W_{jl}
\end{align*}
Stokes
\begin{eqnarray*}
\nabla^{2}\mathbf{u} - \boldsymbol{\nabla}\mathrm{P} = -\alpha_{0}\boldsymbol{\nabla}\cdot\boldsymbol{\Sigma}\
\boldsymbol{\nabla}\cdot\mathbf{u}=\mathbf{0}
\end{eqnarray*}
On the surface or boundary of the disk all fluxes are zero, and the velocity is zero. and the solver im using for all the equations are this:
for , rho, m and Q
problemr = fem.petsc.LinearProblem(ar, Lr,
petsc_options={“ksp_type”: “gmres”,
“pc_type”: “hypre”})
uh = problemr.solve()
uh.x.scatter_forward()
and for velocity of the flow:
solver = fem.petsc.LinearProblem(
a, L, bcs, petsc_options={“ksp_type”: “preonly”, “pc_type”: “lu”,
“pc_factor_mat_solver_type”: “mumps”})
w_h = solver.solve()
bcs this is the boundary dirircthlet conditions.
and this is my meshing:
– Geometría y mallado sólo en rank 0 (serial)
if rank == 0:
gmsh.initialize()
gmsh.model.add(“circle”)
circle = gmsh.model.occ.addDisk(0, 0, 0, radius, radius)
gmsh.model.occ.synchronize()
gmsh.model.addPhysicalGroup(2, [circle], tag=1)
gmsh.model.setPhysicalName(2, 1, “domain”)
boundary = gmsh.model.getBoundary([(2, circle)], oriented=False, recursive=False)
curve_tags = [b[1] for b in boundary if b[0] == 1]
gmsh.model.addPhysicalGroup(1, curve_tags, tag=2)
gmsh.model.setPhysicalName(1, 2, “boundary”)
gmsh.option.setNumber(“Mesh.CharacteristicLengthMax”, h_min)
gmsh.model.mesh.generate(2)
– Colectivo: crea malla distribuida en DOLFINx, todos los ranks participan
domain, cell_tags, facet_tags = gmshio.model_to_mesh(gmsh.model, comm, 0, gdim=2)
if rank == 0:
gmsh.finalize()