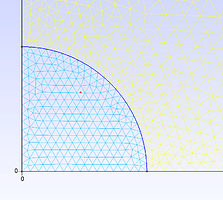
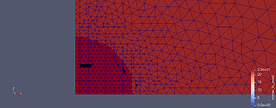
Dear everyone,
I generated mesh with gmsh and it looks normally. Then the mesh is converted into xdmf file and the mesh is messy as it’s viewed with paraview. The figures are the mesh looks in Gmsh editor and Paraview respectively. I don’t know it’s the problem with paraview or the transform code.
The transformation code is:
def create_mesh(mesh, cell_type, prune_z=False):
cells = mesh.get_cells_type(cell_type)
cell_data = mesh.get_cell_data("gmsh:physical", cell_type)
out_mesh = meshio.Mesh(points=mesh.points, cells={cell_type: cells}, cell_data={"name_to_read":[cell_data]})
if prune_z:
out_mesh.prune_z_0()
return out_mesh
msh = meshio.read("mesh.msh")
# Create and save one file for the mesh, and one file for the facets
triangle_mesh = create_mesh(msh, "triangle", prune_z=True)
line_mesh = create_mesh(msh, "line", prune_z=True)
meshio.write("mesh.xdmf", triangle_mesh)
meshio.write("mt.xdmf", line_mesh)
The gmsh file used to generate mesh is :
// Gmsh project created on Tue Oct 19 20:46:26 2021
//+
Point(1) = {0, 0, 0, 0.0001};
//+
Point(2) = {0.0015, 0, 0, 0.0001};
//+
Point(3) = {0, 0.0015, 0, 0.0001};
//+
Point(4) = {0, 0.015, 0, 0.001};
//+
Point(5) = {0.015, 0, 0, 0.001};
//+
Point(6) = {0.15, 0, 0, 0.01};
//+
Point(7) = {0, 0.15, 0, 0.01};
//+
Point(8) = {0, 1.5, 0, 0.1};
//+
Point(9) = {1.5, 0, 0, 0.1};
//+
Point(10) = {15, 0, 0, 1};
//+
Point(11) = {0, 15, 0, 1};
//+
Point(12) = {0, 150, 0, 10};
//+
Point(13) = {150, 0, 0, 10};
//+
Point(14) = {1500, 0, 0, 100};
//+
Point(15) = {0, 1500, 0, 100};
//+
Line(1) = {14, 13};
//+
Line(2) = {15, 12};
//+
Line(3) = {13, 10};
//+
Line(4) = {12, 11};
//+
Line(5) = {10, 9};
//+
Line(6) = {11, 8};
//+
Line(7) = {9, 6};
//+
Line(8) = {8, 7};
//+
Line(9) = {6, 5};
//+
Line(10) = {7, 4};
//+
Line(11) = {5, 2};
//+
Line(12) = {4, 3};
//+
Line(13) = {2, 1};
//+
Line(14) = {3, 1};
//+
Circle(15) = {2, 1, 3};
//+
Circle(16) = {5, 1, 4};
//+
Circle(17) = {6, 1, 7};
//+
Circle(18) = {9, 1, 8};
//+
Circle(19) = {10, 1, 11};
//+
Circle(20) = {13, 1, 12};
//+
Circle(21) = {14, 1, 15};
//+
Curve Loop(1) = {21, 2, -20, -1};
//+
Plane Surface(1) = {1};
//+
Curve Loop(2) = {20, 4, -19, -3};
//+
Plane Surface(2) = {2};
//+
Curve Loop(3) = {19, 6, -18, -5};
//+
Plane Surface(3) = {3};
//+
Curve Loop(4) = {18, 8, -17, -7};
//+
Plane Surface(4) = {4};
//+
Curve Loop(5) = {17, 10, -16, -9};
//+
Plane Surface(5) = {5};
//+
Curve Loop(6) = {16, 12, -15, -11};
//+
Plane Surface(6) = {6};
//+
Curve Loop(7) = {15, 14, -13};
//+
Plane Surface(7) = {7};
//+
Physical Surface("Air", 22) = {1, 2, 3, 4, 5, 6};
//+
Physical Surface("Conductor", 23) = {7};
//+
Physical Curve("Boundary", 24) = {20};